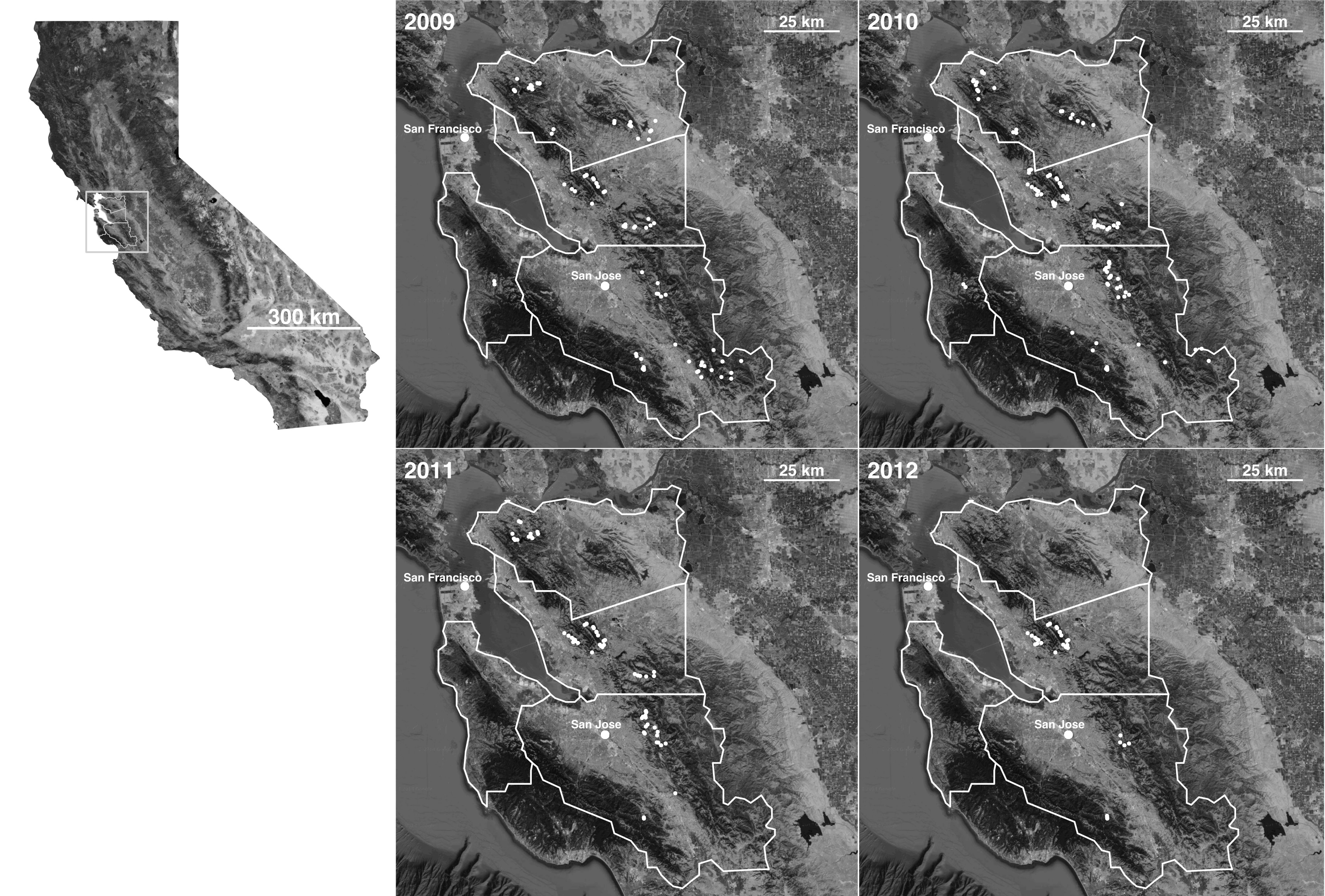
Symbiont Metacommunities
By understanding the ultimate drivers of host and pathogen diversity across space, we should gain a more holistic understanding of infectious disease dynamics in wildlife systems. Community ecology, and in particular metacommunity ecology offers mathematical theory and statistical tools to study the local and regional processes that contribute to coexistence within and among habitats.
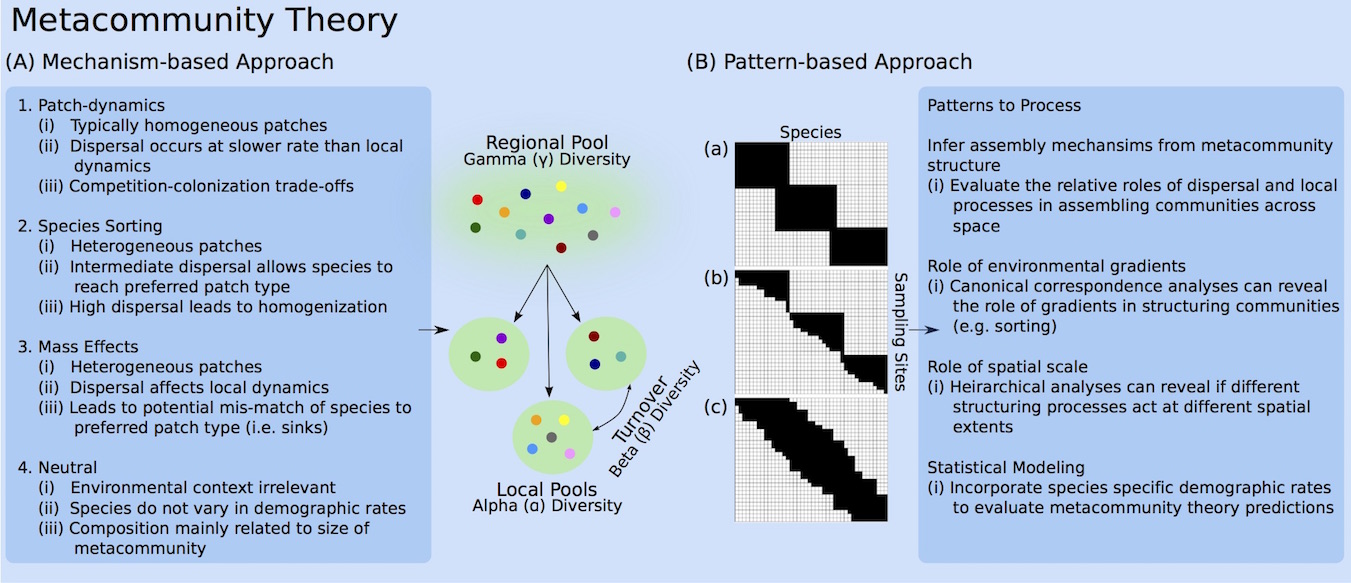
In the metacommunity framework, species communities reside in discrete habitat patches that are distributed across the landscape. Species’ population dynamics, then, are affected both by local processes, such as competition, and by regional processes, such as migration and gene flow. In the case of host-symbiont relationships, host individuals could be seen as patches in (or on) which symbionts reside. Despite the analogous conceptualization of a metacommunity, however, until very recently the metacommunity framework had not been applied symbiotic organisms, likely due to difficulties in sampling microscopic organisms (e.g. Mihaljevic 2012). With the recent emergence of inexpensive, high-throughput sequencing techniques, these sampling barriers have crumbled and a vast trove of symbiont community survey data exists. Thus, there is a great opportunity to (1) use symbiont communities to test basic ecological theory, and (2) to develop theory around host-symbiont dynamics in order to gain a deeper understanding of the mechanisms that shape host and symbiont community composition across space. I am particularly interested in combining models, data, and experiments to determine the relative influences of ecological and epidemiological processes in structuring pathogen communities within and among hosts, and then scaling up to among host habitats.